Basics of R Programming
Thiyanga S. Talagala, University of Sri Jayewardenepura
IASSL - Feb 21/25, 2022
Data structures
Way to store and organize data so that it can be used efficiently.
marks <- c(100, 40, 34, 97, 98)marks[1] 100 40 34 97 98Data structures
Way to store and organize data so that it can be used efficiently.
marks <- c(100, 40, 34, 97, 98)marks[1] 100 40 34 97 98Functions
Tell R to do something
mean(marks)[1] 73.8summary(marks) Min. 1st Qu. Median Mean 3rd Qu. Max. 34.0 40.0 97.0 73.8 98.0 100.0Data structures
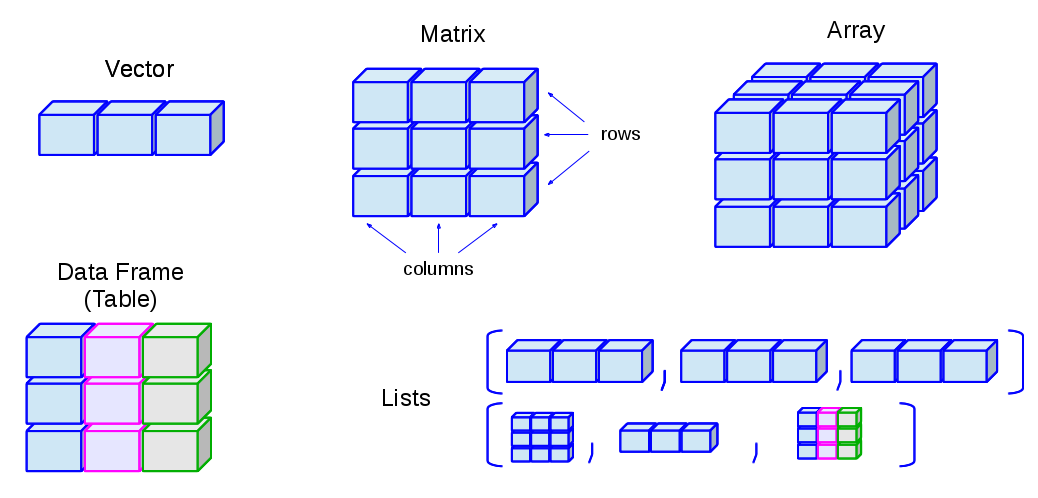
Source: Ceballos and Cardiel, 2013
Creating vectors
Syntax
vector_name <- c(element1, element2, element3)Example
x <- c(5, 6, 3, 1, 100)x[1] 5 6 3 1 100Combine two vectors
p <- c(1, 2, 3)p[1] 1 2 3q <- c(10, 20, 30)q[1] 10 20 30r <- c(p, q)r[1] 1 2 3 10 20 30Vector with charactor elements
names <- c("USJ", "UM", "UC", "UJ")names[1] "USJ" "UM" "UC" "UJ"Logical vector
result <- c(TRUE, FALSE, FALSE, TRUE, FALSE)result[1] TRUE FALSE FALSE TRUE FALSESimplifying vector creation
id <- 1:10id [1] 1 2 3 4 5 6 7 8 9 10treatment <- rep(1:3, each=2)treatment[1] 1 1 2 2 3 3Additional resources: https://hellor.netlify.app/2021/week1/l12021.html#62
Vector operations
x <- c(1, 2, 3)y <- c(10, 20, 30)x+y[1] 11 22 33p <- c(100, 1000)x+p[1] 101 1002 103Your turn
Generate a sequence using the code
seq(from=1, to=10, by=1).What other ways can you generate the same sequence?
Using the function
rep, create the below sequence 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4
03:00
Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[1][1] 1Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[1][1] 1myvec[5:10][1] 5 6 7 8 9 10Vectors: Subsetting (cont.)
Vectors: Subsetting (cont.)
myvec[-1] [1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Vectors: Subsetting (cont.)
myvec[-1] [1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[myvec > 3] [1] 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Changing values of a vector
covid <- c(100, 30, 40, 50, -1, 100)covid[1] 100 30 40 50 -1 100covid[1] <- 50000covid[1] 50000 30 40 50 -1 100Changing values of a vector (cont.)
covid[covid < 0] <- 0covid[1] 50000 30 40 50 0 100covid[c(1, 2)] <- c(1000, 10000)covid[1] 1000 10000 40 50 0 100factor
Required R package
library(tidyverse)── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──✓ ggplot2 3.3.5 ✓ purrr 0.3.4 ✓ tibble 3.1.6 ✓ dplyr 1.0.8.9000✓ tidyr 1.2.0 ✓ stringr 1.4.0 ✓ readr 2.1.2 ✓ forcats 0.5.1Warning: package 'tidyr' was built under R version 4.1.2Warning: package 'readr' was built under R version 4.1.2── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──x dplyr::filter() masks stats::filter()x dplyr::lag() masks stats::lag()Character vector vs Factor
- Factor prints all possible levels of the variable.
Character vector
grade_character_vctr <- c("A", "D", "A", "C", "B")grade_character_vctr[1] "A" "D" "A" "C" "B"Factor vector
grade_factor_vctr <- factor(c("A", "D", "A", "C", "B"), levels = c("A", "B", "C", "D", "E"))grade_factor_vctr[1] A D A C BLevels: A B C D ECharacter vector vs Factor (cont.)
- Let's create a contingency table with
tablefunction.
Character vector output with table function
grade_character_vctr <- c("A", "D", "A", "C", "B")table(grade_character_vctr)grade_character_vctrA B C D 2 1 1 1Factor vector (with levels) output with table function
grade_factor_vctr <- factor(c("A", "D", "A", "C", "B"), levels = c("A", "B", "C", "D", "E"))table(grade_factor_vctr)grade_factor_vctrA B C D E 2 1 1 1 0- Output corresponds to factor prints counts for all possible levels of the variable. Hence, with factors it is obvious when some levels contain no observations.
Character vector vs Factor (cont.)
- With factors you can't use values that are not listed in the levels, but with character vectors there is no such restrictions.
Character vector
grade_character_vctr[2] <- "A+"grade_character_vctr[1] "A" "A+" "A" "C" "B"Factor vector
grade_factor_vctr[2] <- "A+"grade_factor_vctr[1] A <NA> A C B Levels: A B C D EFactor: order levels
fv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 1T 2T 3A 4A 5A 6BFactor: order levels
fv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 1T 2T 3A 4A 5A 6Blibrary(ggplot2)qplot(fv2, geom = "bar")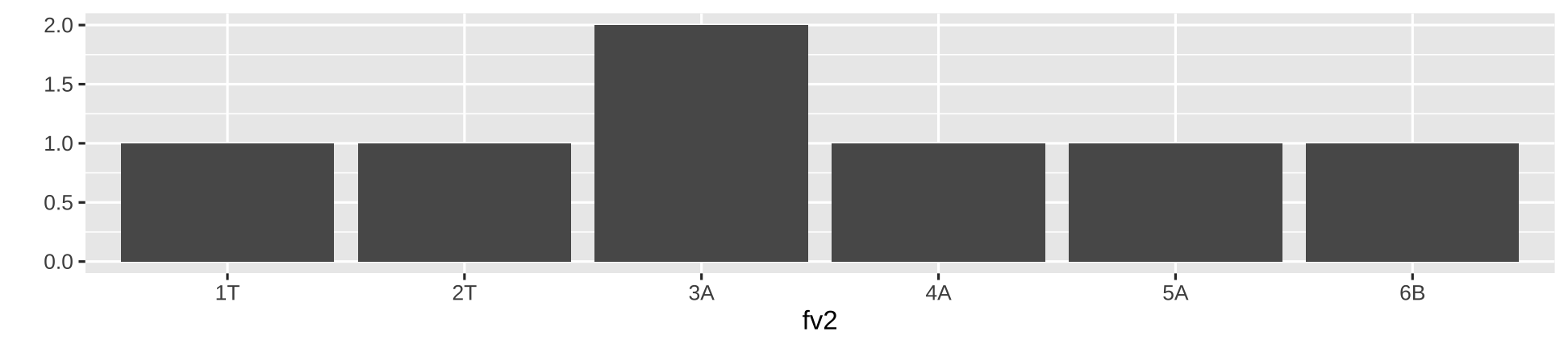
You can change the order of levels
fv2 <- factor(c("1T","2T","3A","4A", "5A", "6B", "3A"), levels = c("3A", "4A", "5A", "6B", "1T", "2T"))fv2[1] 1T 2T 3A 4A 5A 6B 3ALevels: 3A 4A 5A 6B 1T 2Tqplot(fv2, geom = "bar")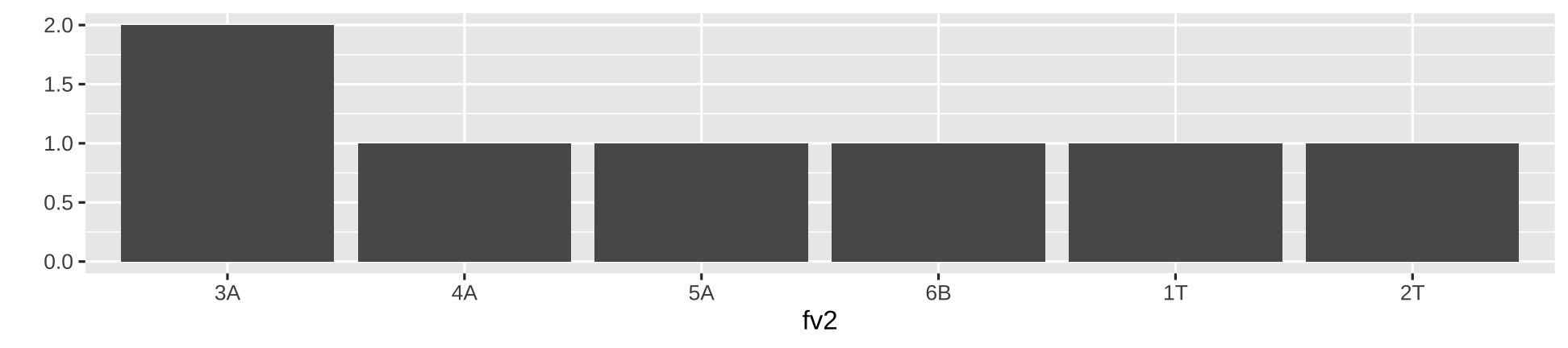
Data set
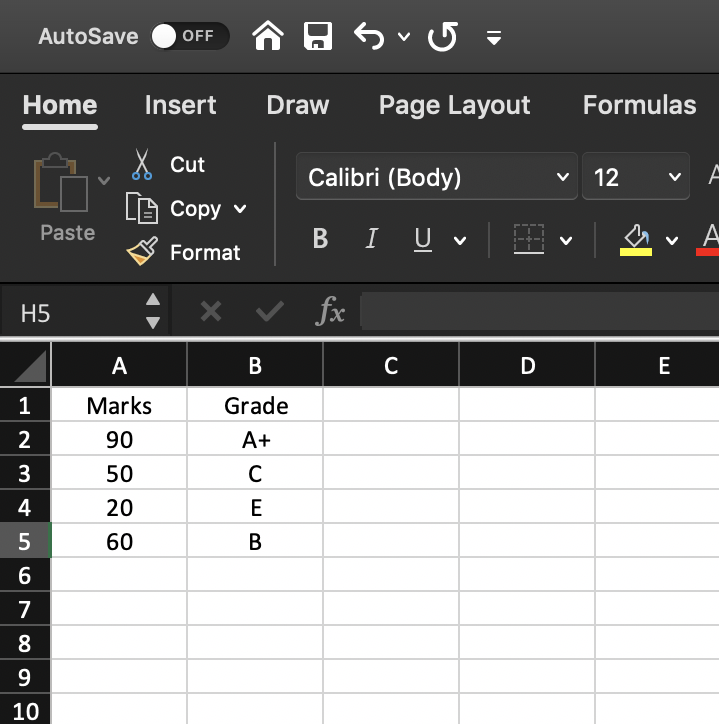
Required R package
library(tidyverse)Create a tibble

marks <- c(90, 50, 20, 60)grade <- factor(c("A+", "C", "E", "B"))final <- tibble(Marks = marks, Grade = grade)final# A tibble: 4 × 2 Marks Grade <dbl> <fct>1 90 A+ 2 50 C 3 20 E 4 60 BCreate a tibble
marks <- c(90, 50, 20, 60)grade <- factor(c("A+", "C", "E", "B"), level = c("A+", "A", "B+", "B", "C", "D", "E"))final <- tibble(Marks = marks, Grade = grade)final# A tibble: 4 × 2 Marks Grade <dbl> <fct>1 90 A+ 2 50 C 3 20 E 4 60 BFunctions in R
Data set: tibble
final# A tibble: 4 × 2 Marks Grade <dbl> <fct>1 90 A+ 2 50 C 3 20 E 4 60 BFunctions
summary(final) Marks Grade Min. :20.0 A+:1 1st Qu.:42.5 A :0 Median :55.0 B+:0 Mean :55.0 B :1 3rd Qu.:67.5 C :1 Max. :90.0 D :0 E :1Your Turn
01:00

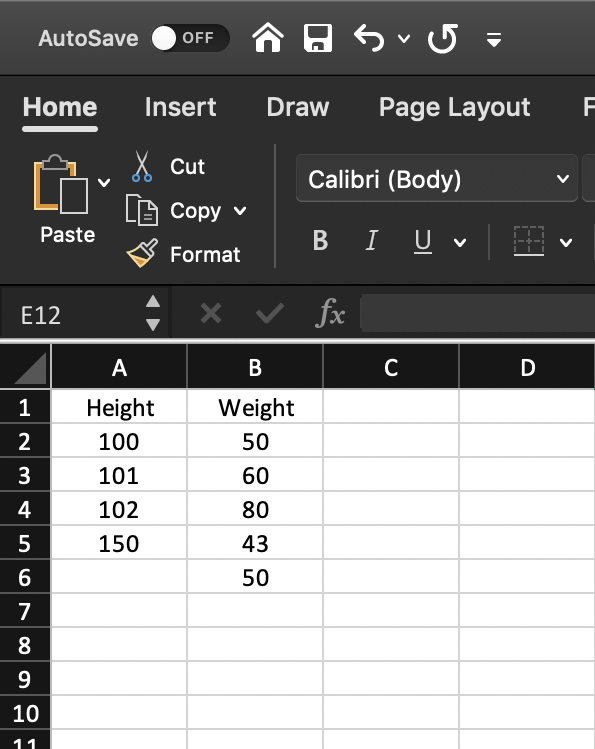
h <- c(100, 101, 102, 150, NA)w <- c(50, 60, 80, 43, 50)hwdata <- tibble(Height=h, Weight=w)hwdata# A tibble: 5 × 2 Height Weight <dbl> <dbl>1 100 502 101 603 102 804 150 435 NA 50hwdata# A tibble: 5 × 2 Height Weight <dbl> <dbl>1 100 502 101 603 102 804 150 435 NA 50summary(hwdata) Height Weight Min. :100.0 Min. :43.0 1st Qu.:100.8 1st Qu.:50.0 Median :101.5 Median :50.0 Mean :113.2 Mean :56.6 3rd Qu.:114.0 3rd Qu.:60.0 Max. :150.0 Max. :80.0 NA's :1Subsetting
hwdata# A tibble: 5 × 2 Height Weight <dbl> <dbl>1 100 502 101 603 102 804 150 435 NA 50hwdata[1, 1]# A tibble: 1 × 1 Height <dbl>1 100hwdata[, 1]# A tibble: 5 × 1 Height <dbl>1 1002 1013 1024 1505 NAhwdata[1, ]# A tibble: 1 × 2 Height Weight <dbl> <dbl>1 100 50hwdata$Height[1] 100 101 102 150 NAHelp file
hwdata$Weight[1] 50 60 80 43 50mean(hwdata$Weight)[1] 56.6hwdata$Height[1] 100 101 102 150 NAmean(hwdata$Height)[1] NAHelp file
hwdata$Weight[1] 50 60 80 43 50mean(hwdata$Weight)[1] 56.6hwdata$Height[1] 100 101 102 150 NAmean(hwdata$Height)[1] NAmean(hwdata$Height, na.rm=TRUE)[1] 113.25Help file
?meanhelp(mean)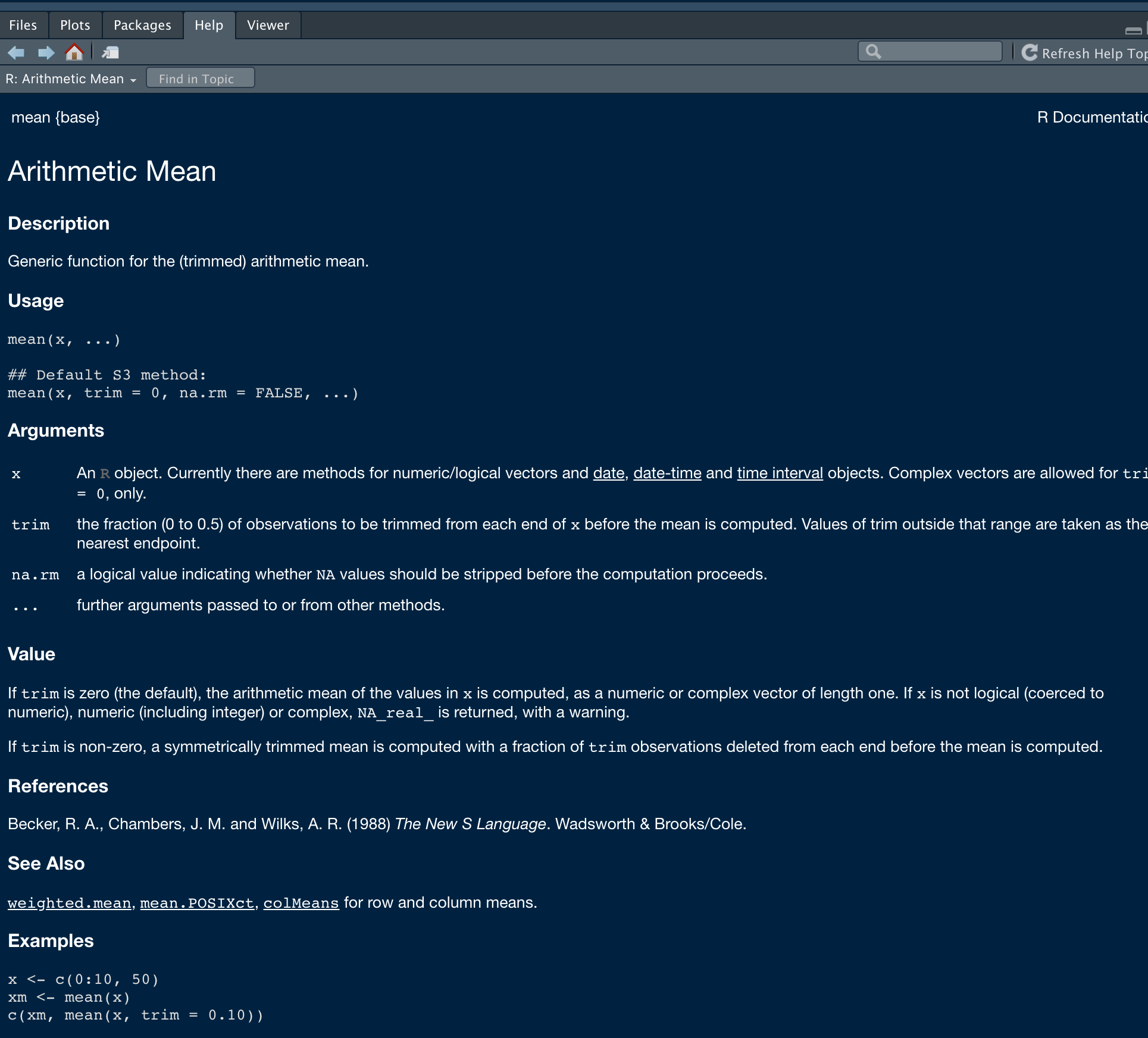
Commenting
mean(hwdata$Height, na.rm=TRUE) # compute mean of height[1] 113.25Some useful functions
mean(hwdata$Weight)[1] 56.6median(hwdata$Weight)[1] 50sd(hwdata$Weight)[1] 14.41527sum(hwdata$Weight)[1] 283length(hwdata$Weight)[1] 5Pipe operator (%>%)
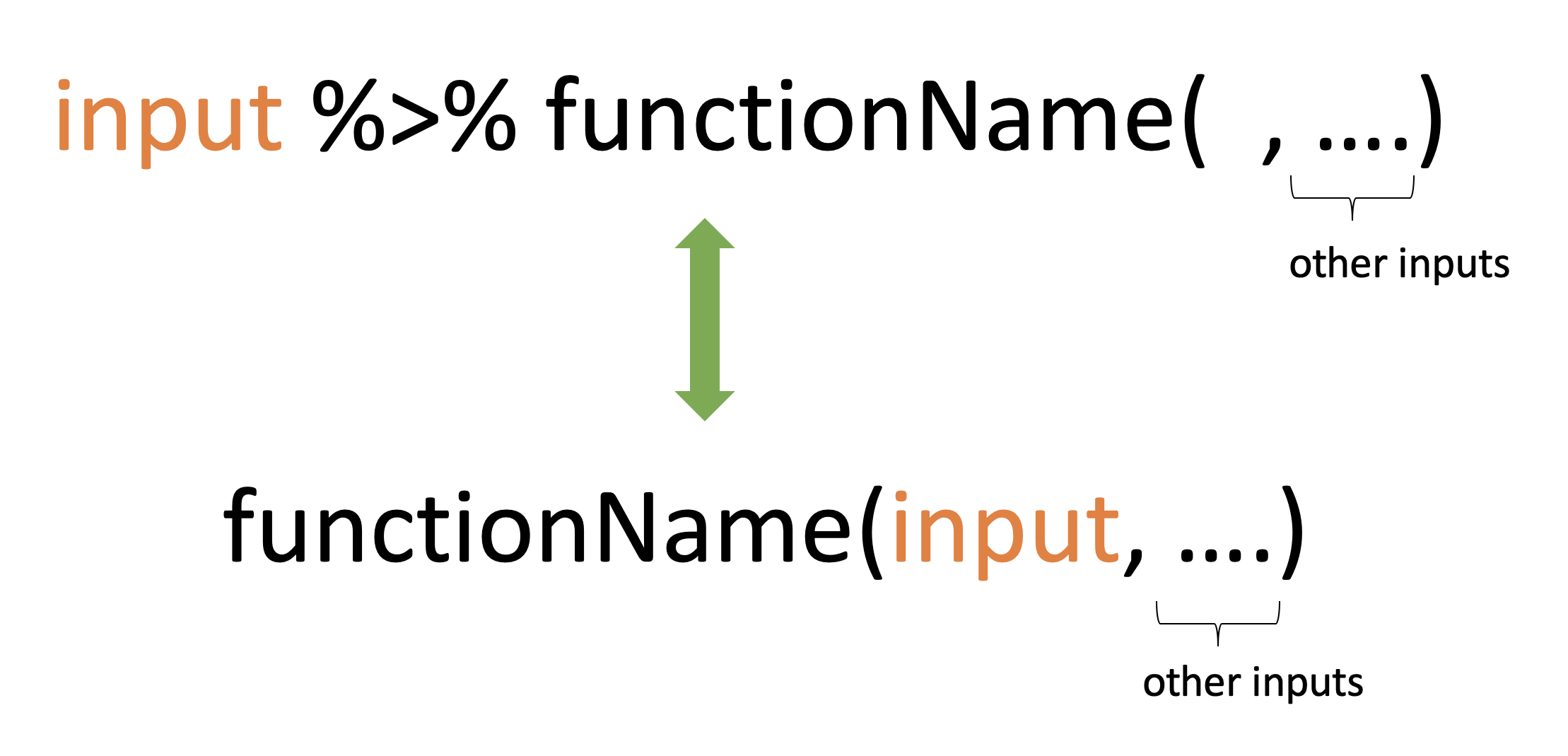
mean(hwdata$Weight)[1] 56.6mean(hwdata$Height, na.rm=TRUE)[1] 113.25library(magrittr)hwdata$Weight %>% mean()[1] 56.6hwdata$Height %>% mean(na.rm=TRUE)[1] 113.25Pipe operator (%>%)
Built-in dataset
library(palmerpenguins)data(penguins)head(penguins)# A tibble: 6 × 8 species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex <fct> <fct> <dbl> <dbl> <int> <int> <fct>1 Adelie Torge… 39.1 18.7 181 3750 male 2 Adelie Torge… 39.5 17.4 186 3800 fema…3 Adelie Torge… 40.3 18 195 3250 fema…4 Adelie Torge… NA NA NA NA <NA> 5 Adelie Torge… 36.7 19.3 193 3450 fema…6 Adelie Torge… 39.3 20.6 190 3650 male # … with 1 more variable: year <int>Skim data
library(skimr)skim(penguins)iris dataset

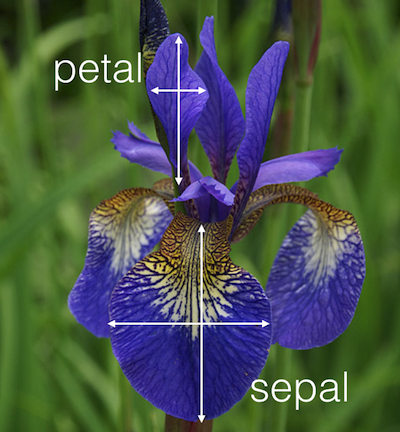
Use the R dataset “iris” to answer the following questions:
How many rows and columns does iris have?
Select the first 4 rows.
Select the last 6 rows.
Select rows 10 to 20, with all columns in the iris dataset.
Select rows 10 to 20 with only the Species, Petal.Width and Petal.Length.
Create a single vector (a new object) called ‘width’ that is the Sepal.Width column of iris.
What are the column names and data types of the different columns in iris?
How many rows in the iris dataset have
Petal.Lengthlarger than 5 andSepal.Widthsmaller than 3?
05:00
Recap
✅ Data structures and functions
✅ Factors
✅ Working with packages
✅ Create a tibble
✅ Help file
✅ Commenting